Synthetic Data & Artificial Genomes
Summer 2023 ~ @TitouanCh
In the winter of 2023, I had the pleasure of working with a team on artificial genomes generated by neural networks. This is an interesting area very close the synthetic life. This is a very brief overview of the work I've achieved while with this team.
Introduction
In early 2023, I had the opportunity to work as a part-time research assistant on a cool project that involved training neural network models on human genomes. One of the primary challenges in this domain is the limited availability of genomic data and privacy concerns that limit the vailable data to researchers. While initiatives like the Human Genome Project have provided invaluable resources, the quantity of real human genome data remains restricted, which poses significant challenges when training machine learning models or neural networks.
A promising solution to this scarcity of data is the use of synthetic data. This approach offers a way to generate new datasets that retain the key characteristics of real data. It also offers a potential way to address privacy concerns, serving as an intermediary by generating data that reflects real patterns without revealing the original, sensitive information, provided, there's no way to reconstruct the original data from the synthetic data.
Synthetic data
Synthetic data refers to artificially generated data that mimics real-world data. It is produced by generating new data points based on the patterns and statistical properties of an original dataset. The goal is to maintain the essential features of the original data while expanding the dataset to improve the performance of machine learning models.
However, there's some risks to using synthetic data. These risk are mostly related to how you generate the synthetic data but it can lead to overfitting: where models perform well on training data but fail to generalize to real-world scenarios.
Generating synthetic genomes
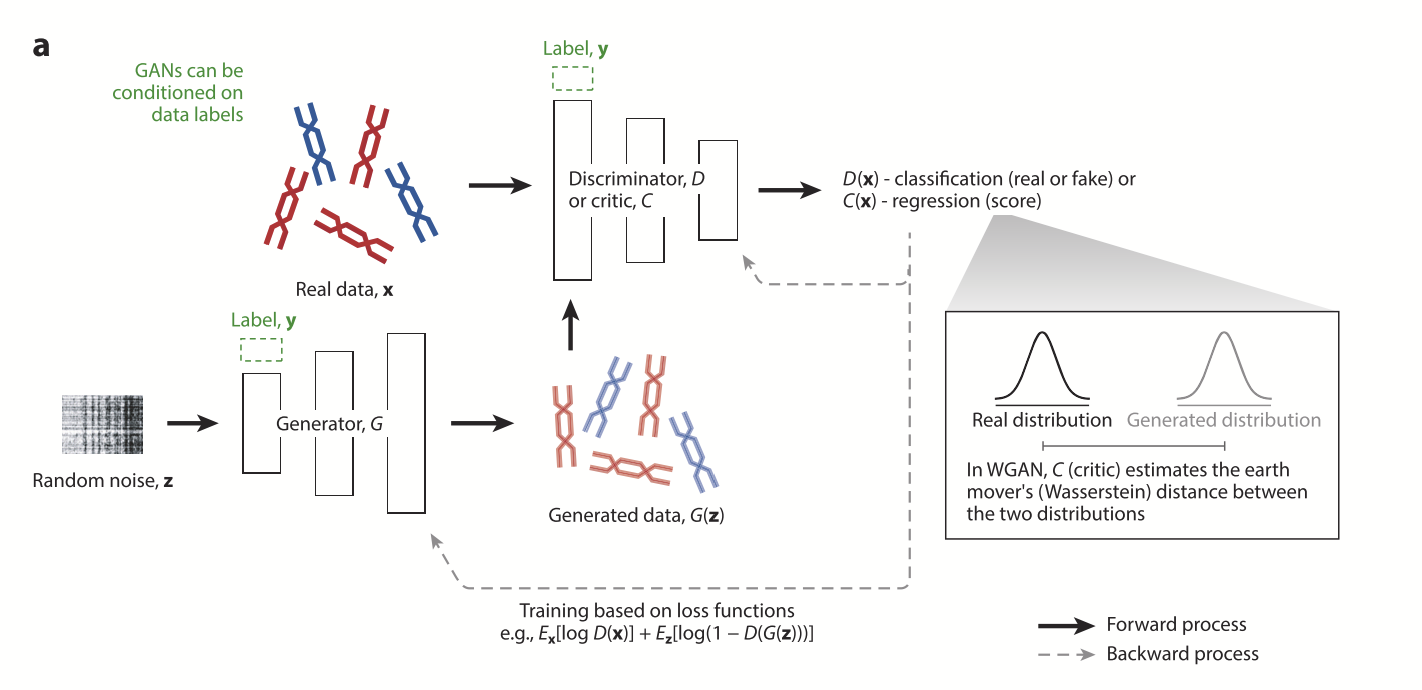
In the project, synthetic genomes were generated using a Wasserstein Generative Adversarial Network (WGAN). WGANs are a specific type of deep learning model designed to generate realistic synthetic data by optimizing a generator and a discriminator in a competitive process. The generator tries to create realistic data, while the discriminator evaluates how similar the generated data is to the real data.
The technical aspects of the WGAN were not the center of my work, so I won't expend too much on it. My focus was on analyzing the synthetic genomic data that was produced. As I've talked about before, the quality of this synthetic data was crucial in determining whether it retained the fundamental properties of the original human genome data and if it was useful or not.
Labeling synthetic data
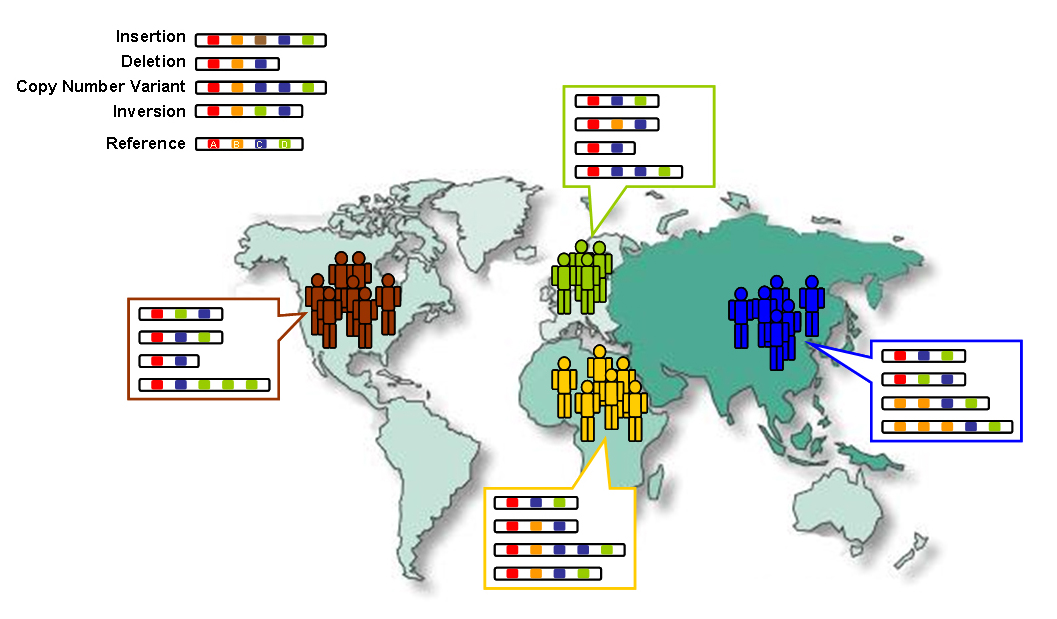
One main topic in population genetics is studying genetic variations among populations. A common task is to predict an individual's geographic ancestry based on their genomic data. For instance, it's possible to determine the continent or even specific regions an individual comes from based solely on their genome and their SNPs (single nucleotide polymorphs).
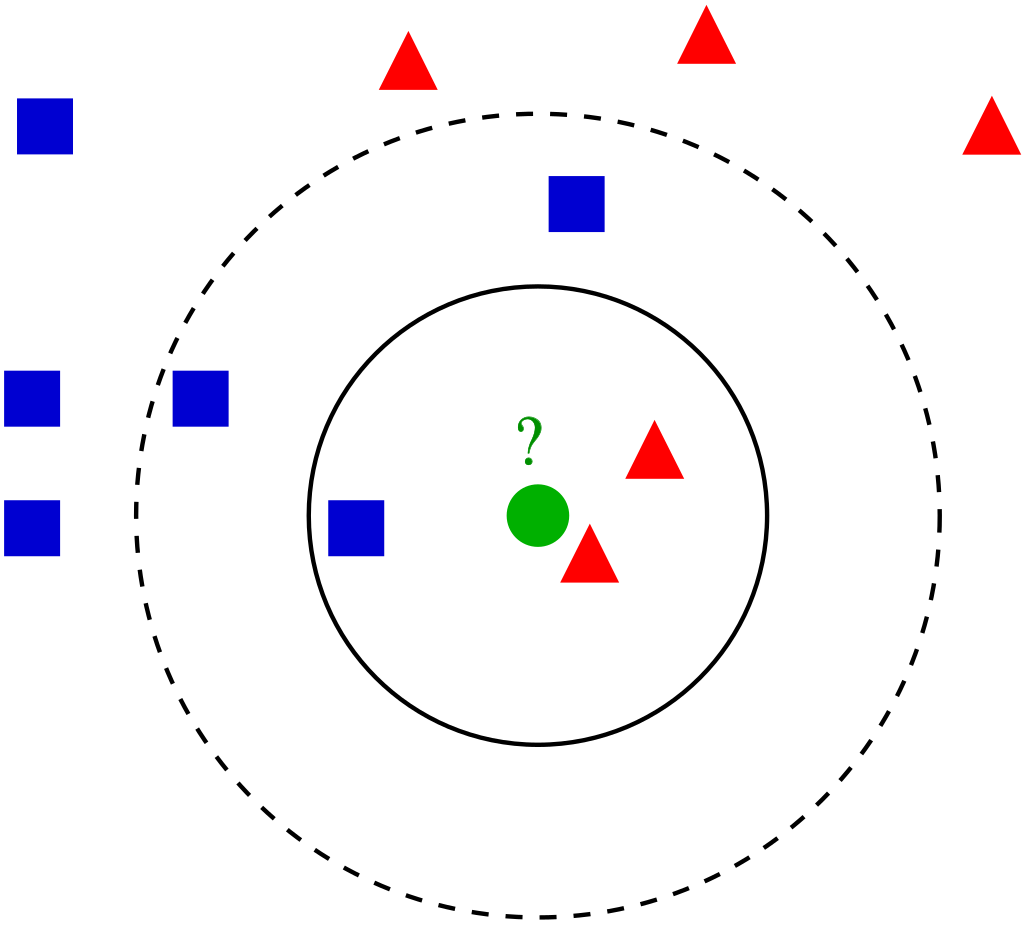
To label our synthetic data, I employed a k-nearest neighbors (KNN) algorithm trained on our orignal real genomic data. KNN is a simple yet effective classification model that assigns labels to data points based on the majority label of its closest neighbors in the dataset.
By using real genomic data, we could train the KNN to classify synthetic genomes according to their probable population group. We also ensured that our synthetic data reflected real-world population distributions.
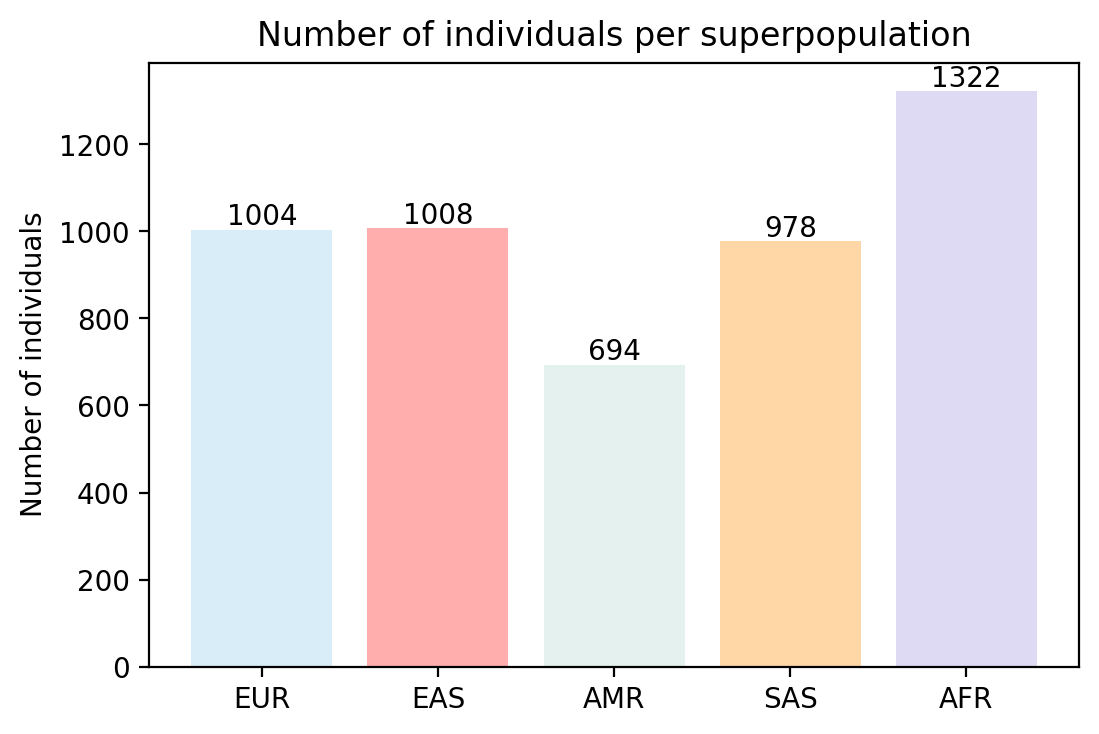
Bar graph showing the different proportion of haplotype of each superpopulation in the real data
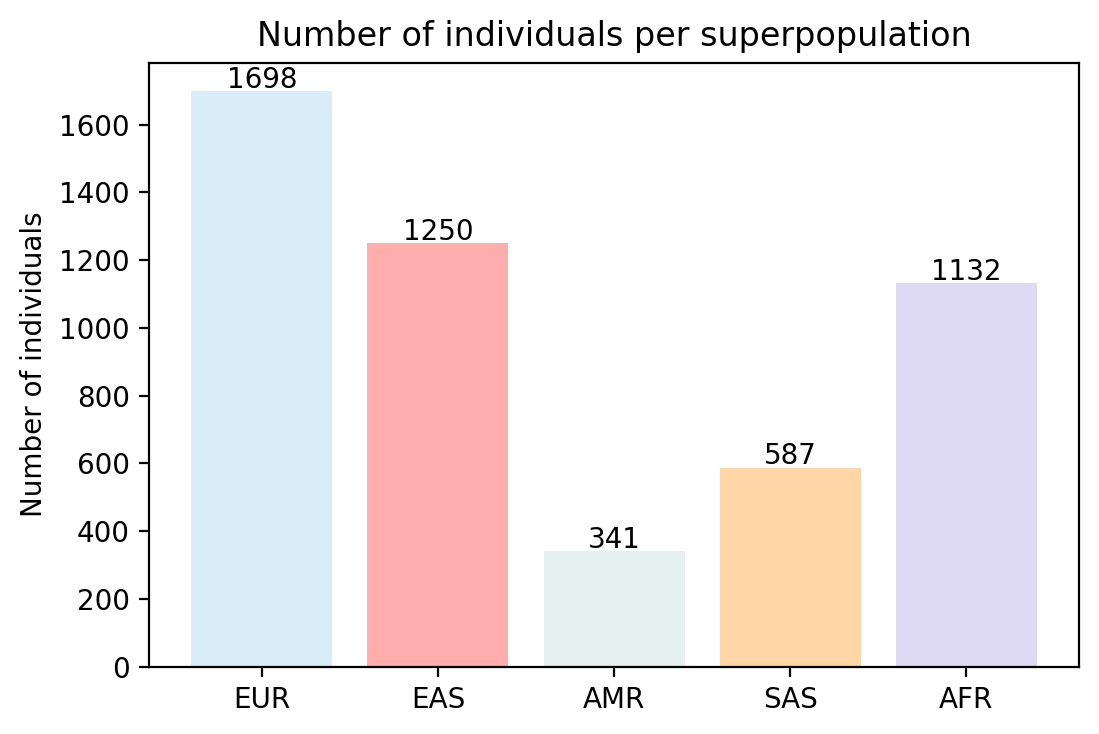
Bar graph showing the different proportion of haplotype of each superpopulation in the synthetic data
It's interesting to note that it seems that some proportions of certain populations were exagerated by our WGAN.
Training neural networks on synthetic data
To assess the quality and utility of the synthetic genomic data, we conducted experiments by training a convolutional neural network (CNN) called LAI-NET on both real and synthetic data. LAI-NET is designed for tasks related to local ancestry inference (LAI), which involves identifying the ancestral origins of different segments of an individual's genome.
The goal was to determine whether the synthetic data was comparable to real data in terms of training effectiveness. If the synthetic data retained the key features of the real data, the performance of the neural network trained on synthetic data should be similar to that trained on real data.
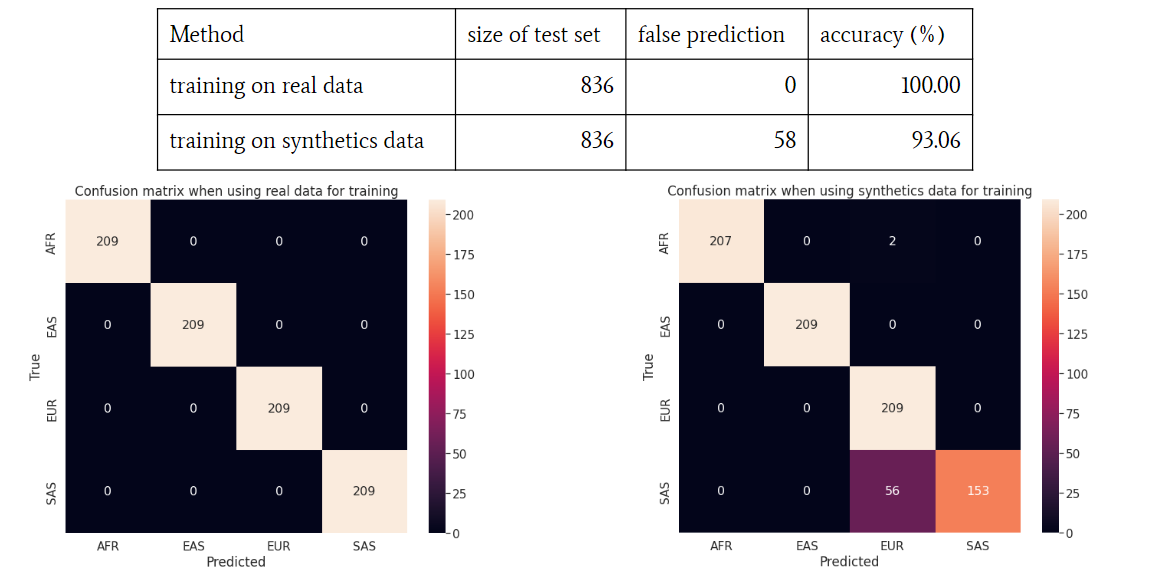
Results
Our results were encouraging. LAI-NET achieved comparable performance regardless of whether it was trained on real or synthetic genomic data. This finding suggested that the synthetic data successfully preserved the essential features of the real genomic data, making it a valuable tool for future research in genomics and AI.
Conclusion
The use of synthetic data in genomics opens up new possibilities for advancing research and training machine learning models, especially in areas where data is scarce or limited due to privacy concerns. By generating high-quality synthetic genomes, we can enhance AI models' ability to learn from genetic data without compromising on accuracy. However, as with any tool, synthetic data must be used responsibly to avoid overfitting and ensure that models trained on synthetic data can generalize well to real-world applications.